Predicting Patient Outcomes with Recurrent Deep Learning and Attention
PYTORCH
PYSPARK
SCIKIT-LEARN
This project focuses on reproducing ConCare [1], a GRU-based deep learning model with multiple attention mechanisms, designed for ICU mortality prediction using irregular and heterogeneous clinical time-series data from the MIMIC-III healthcare database. Unlike previous modeling approaches that compress each patient’s history into a single sequence, ConCare models each clinical variable independently, allowing the network to learn how different vital signs and lab measurements influence risk over time.
This project demonstrated the strengths of ConCare’s feature-level modeling and showed that its personalized, temporally aware architecture could be successfully reproduced with comparable performance. The reproduced model and extension experiments provided additional insight into how time-aware attention, demographic context, and feature-level interactions contributed to effective clinical risk prediction.
Ref: [1] ConCare: Personalized Clinical Feature Embedding via Capturing the Healthcare Context, Liantao Ma, Tianyu Zhang, Yuntao Wang, et al. AAAI 2020


The reproduction began by rebuilding the dataset pipeline and preparing both the dynamic clinical measurements and static demographic information required for training. The data was extracted and structured from the raw MIMIC-III tables to create aligned episode-level inputs for the model. The training environment was implemented in modern PyTorch (2.5) with CUDA support, enabling efficient computation on current GPU hardware.
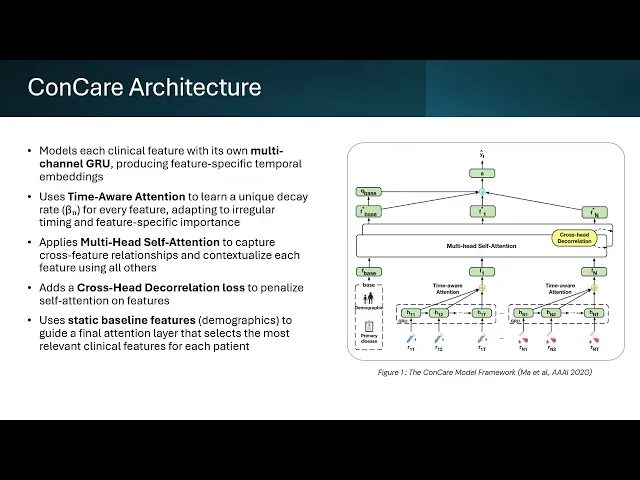
The model architecture mirrored the original design: per-feature GRUs extract temporal embeddings, time-aware attention learns a feature-specific decay rate to handle irregular sampling, and multi-head self-attention captures cross-feature relationships. A decorrelation loss encourages diverse attention heads, and demographic variables guide a final attention layer to produce a personalized patient representation.
Our reproduced model achieved performance close to the paper’s reported benchmarks, confirming that ConCare’s main ideas remain effective under updated implementations. Model performance evaluation was primarily based on the AUROC and AUPRC metrics, aligning with the original study’s focus on imbalanced clinical outcomes. We also explored several extensions, including Asymmetric Focal BCE to improve recall on rare mortality cases and temporal masking regularization to simulate missing data. These techniques enhanced sensitivity and robustness, highlighting practical ways to adapt clinical prediction models to real-world EMR conditions.


This project focuses on reproducing ConCare [1], a GRU-based deep learning model with multiple attention mechanisms, designed for ICU mortality prediction using irregular and heterogeneous clinical time-series data from the MIMIC-III healthcare database. Unlike previous modeling approaches that compress each patient’s history into a single sequence, ConCare models each clinical variable independently, allowing the network to learn how different vital signs and lab measurements influence risk over time.

The reproduction began by rebuilding the dataset pipeline and preparing both the dynamic clinical measurements and static demographic information required for training. The data was extracted and structured from the raw MIMIC-III tables to create aligned episode-level inputs for the model. The training environment was implemented in modern PyTorch (2.5) with CUDA support, enabling efficient computation on current GPU hardware.
The model architecture mirrored the original design: per-feature GRUs extract temporal embeddings, time-aware attention learns a feature-specific decay rate to handle irregular sampling, and multi-head self-attention captures cross-feature relationships. A decorrelation loss encourages diverse attention heads, and demographic variables guide a final attention layer to produce a personalized patient representation.
Our reproduced model achieved performance close to the paper’s reported benchmarks, confirming that ConCare’s main ideas remain effective under updated implementations. Model performance evaluation was primarily based on the AUROC and AUPRC metrics, aligning with the original study’s focus on imbalanced clinical outcomes. We also explored several extensions, including Asymmetric Focal BCE to improve recall on rare mortality cases and temporal masking regularization to simulate missing data. These techniques enhanced sensitivity and robustness, highlighting practical ways to adapt clinical prediction models to real-world EMR conditions.
This project demonstrated the strengths of ConCare’s feature-level modeling and showed that its personalized, temporally aware architecture could be successfully reproduced with comparable performance. The reproduced model and extension experiments provided additional insight into how time-aware attention, demographic context, and feature-level interactions contributed to effective clinical risk prediction.
Ref: [1] ConCare: Personalized Clinical Feature Embedding via Capturing the Healthcare Context, Liantao Ma, Tianyu Zhang, Yuntao Wang, et al. AAAI 2020


The reproduction began by rebuilding the dataset pipeline and preparing both the dynamic clinical measurements and static demographic information required for training. The data was extracted and structured from the raw MIMIC-III tables to create aligned episode-level inputs for the model. The training environment was implemented in modern PyTorch (2.5) with CUDA support, enabling efficient computation on current GPU hardware.
The model architecture mirrored the original design: per-feature GRUs extract temporal embeddings, time-aware attention learns a feature-specific decay rate to handle irregular sampling, and multi-head self-attention captures cross-feature relationships. A decorrelation loss encourages diverse attention heads, and demographic variables guide a final attention layer to produce a personalized patient representation.
Our reproduced model achieved performance close to the paper’s reported benchmarks, confirming that ConCare’s main ideas remain effective under updated implementations. Model performance evaluation was primarily based on the AUROC and AUPRC metrics, aligning with the original study’s focus on imbalanced clinical outcomes. We also explored several extensions, including Asymmetric Focal BCE to improve recall on rare mortality cases and temporal masking regularization to simulate missing data. These techniques enhanced sensitivity and robustness, highlighting practical ways to adapt clinical prediction models to real-world EMR conditions.


This project focuses on reproducing ConCare [1], a GRU-based deep learning model with multiple attention mechanisms, designed for ICU mortality prediction using irregular and heterogeneous clinical time-series data from the MIMIC-III healthcare database. Unlike previous modeling approaches that compress each patient’s history into a single sequence, ConCare models each clinical variable independently, allowing the network to learn how different vital signs and lab measurements influence risk over time.

This project demonstrated the strengths of ConCare’s feature-level modeling and showed that its personalized, temporally aware architecture could be successfully reproduced with comparable performance. The reproduced model and extension experiments provided additional insight into how time-aware attention, demographic context, and feature-level interactions contributed to effective clinical risk prediction.
Ref: [1] ConCare: Personalized Clinical Feature Embedding via Capturing the Healthcare Context, Liantao Ma, Tianyu Zhang, Yuntao Wang, et al. AAAI 2020

Published December 2025
Go back home